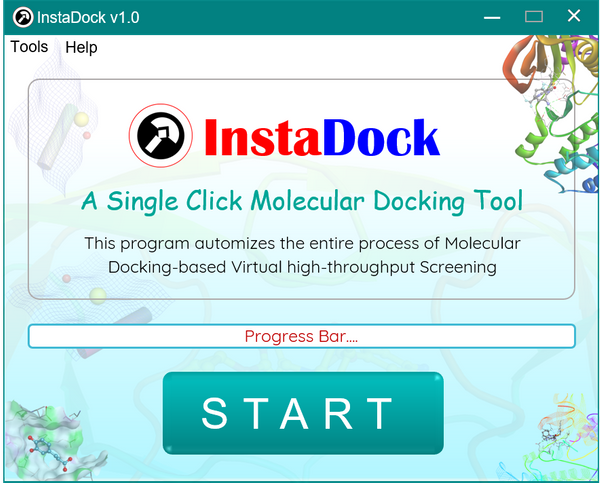
About InstaDock
InstaDock is a front-end graphical user interface written in Python to perform molecular docking-based virtual high-throughput screening that can be done in just one go. It provides users a single-click interactive platform to perform automated continuous docking of large compound databases against predefined protein targets. It will also help the user to visualize and analyze the results to identify promising lead molecules. InstaDock provides a straight forward graphical user interface, and a complete suite to perform molecular docking and high-throughput virtual screening on Windows-based computers.
It employs a few standard programs for docking, molecular conversion and visualization purposes, and several Python scripts for processing and execution. It automatically detects the receptor as well as the ligand molecules in the directory. After the process, it creates a ‘Result’ folder for you containing all docked files, docking scores, top hits, their splitted conformers, and a brief write-up. Various standalone programs available in ‘Tools’ menu of InstaDock gives the user the freedom to perform ‘User-directed Docking’ and different tasks by executing individual programs as described in original publication (Click here).
Please fill this form to start your Download
Please note: Your Download will automatically start in 1-3 seconds after submitting this form. If in case your Windows Defender alerts you for InstaDock (during your first download/run), just allow it (click -> More info -> Run anyway), the alert is due to a false recognition since the program is new to Windows. Thanks!!
For any queries you can direct write to the following:
Taj (taj@hassanlab.org; taj144796@st.jmi.ac.in)
Yash (yash185973@st.jmi.ac.in)
Dr. Hassan (hassanlab@jmi.ac.in)
Address for correspondence:
G-04, Srinivasan Ramanujan Block, Centre for Interdisciplinary Research in Basic Sciences, Jamia Millia Islamia, New Delhi-110025, INDIA
How to Cite
Please cite the following papers while publishing results from InstaDock:
- Mohammad T, Mathur Y, Hassan MI (2021). InstaDock: A Single-click Graphical User Interface for Molecular Docking-based Virtual High-throughput Screening. Briefings in Bioinformatics, 22(4), bbaa279, https://doi.org/10.1093/bib/bbaa279.
- Hassan NM, Alhossary AA, Mu Y, Kwoh CK (2017). Protein-ligand blind docking using QuickVina-W with inter-process spatio-temporal integration, Scientific reports, 7(1): 1-13.
You are also encouraged to cite the following references where credit is due:
- Trott O, Olson AJ (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of computational chemistry, 31(2): 455-461.
- O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011). Open Babel: An open chemical toolbox. Journal of cheminformatics, 3(1): 33.
- Sayle RA, Milner-White EJ (1995). RASMOL: biomolecular graphics for all. Trends in biochemical sciences, 20(9): 374-376.
- Bernstein HJ. Recent changes to RasMol, recombining the variants (2000). Trends in biochemical sciences, 25(9): 453-455.
How to use InstaDock
- Place the ‘InstaDock EXE’ file in the same directory as your receptor and ligand files. (The files can be of any standard chemical format i.e. PDB, SDF, MOL, MOL2 or PDBQT).
- Click the ‘START’ button to initiate the automatic process of Molecular Docking. Yes, it is that easy!
Note: The single-click execution performs Blind docking by default. For site-specific OR ‘User-directed docking’, please refer to ‘Tutorials’ published in the original paper available through https://doi.org/10.1093/bib/bbaa279. Happy Docking!! 🙂
Download InstaDock
Please fill this form to start your Download
Video Tutorial
The Team Behind InstaDock
InstaDock development
We are always interested in adding additional functionalities to InstaDock. If you have ideas, suggestions or code that you would like to contribute, please contact us. You are encouraged to share your ideas and suggestions with us at taj144796@st.jmi.ac.in for the future development of InstaDock. Please do get in touch – we would like to hear from you!
Get in touch with Team InstaDock
Drop your message here. We welcome your query, suggestion & valuable feedback-